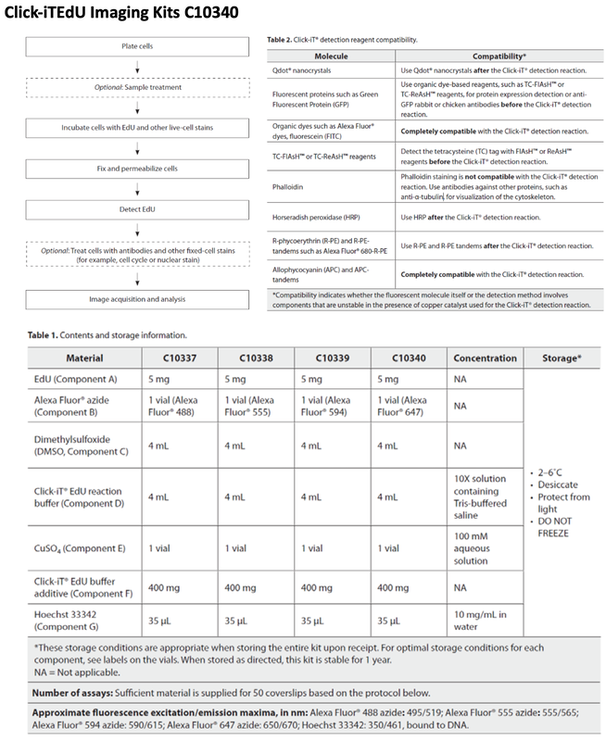
Cell Cycle Control and Chromatin DNA Labelling
MATERIALS
METHODS
Day 0 – Sun May 04 201
16:00 thaw and passage Hela cells from liquid nitrogen.
Day 1 – Mon May 05 2014 11:00 passage cell
In 6‐well plate, passage the Hela cells 1:2 in fresh medium. 19:00 1st Block (15hrs)
In 6‐well plate, change fresh medium.
Add Thymidine to final concentration 2mM to Hela cells medium to block the cells.
Day 2 – Tue May 06 2014 10:00 1st Release (10hrs)
To release the cells from S phase, wash cells with 1X DPBS thrice to remove the high concentration of Thymidine, passage Hela cells to coverslip bottomed petri dish to final density 20% ~ 30%.
#VII‐1 #VII‐2 #VII‐3 Hela cells; plate some cells in 6well plate as #VII‐4 dish;
20:00 2nd Block (15hrs)
In 6‐well plate and 3 coverslip bottomed petri dishes, change fresh medium.
Add Aphidicolin to final concentration 2ug/ml to Hela cell medium
Day 3 - Wed May 07 2014
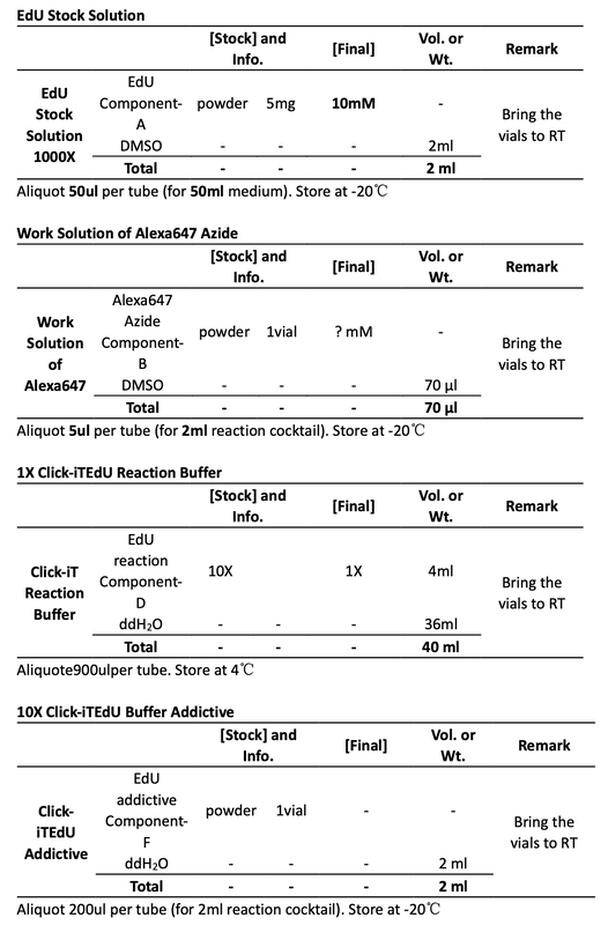
10:20 make 12ml fresh medium containing 10uM EdU in 15ml tube.
Early-S # VI-1dish Mid-S # VI-2 dish Late-S # VI-3dish
10:30 13:00 15:30
100ul PBS + 25ul FuGene6, incubate 5min at RT. Add 1ul 1mM Atto550-dUTP (Jena Bioscience) to the mix, incubate 20min at 4C freezer, final concentration of the Atto550-dUTP is ~10uM.
11:00 # VI-1,# VI-2,# VI-3, # VI-4 dish To release the cells, wash cells with PBS thrice to remove the aphidicolin, change to 4ml fresh medium (DMEM), then the cells will enter S phase.
Whole Chrom # V-4 dish
11:00 3ml fresh medium + 1ml fresh medium with 10uM EdU. Final EdU is 2.5uM.
Early-S # VI-1dish Mid-S # VI-2 dish Late-S # VI-3dish
11:00 13:30 16:00
Wash cells with PBS twice. Add the ~100ul Atto550-dUTP-FuGene6-PBS Mix to PBS washed cells for 10min at RT. Note: be careful, do not let the cell dry!!!
11:10 13:40 16:10
Fill the dish with 4ml FRESH DMEM. Culture cells at 37C for 0.5 hrs.
11:40 14:10 16:40
4ml DMEM medium with EdU, the concentration is 10uM.
12:10 14:40 17:10
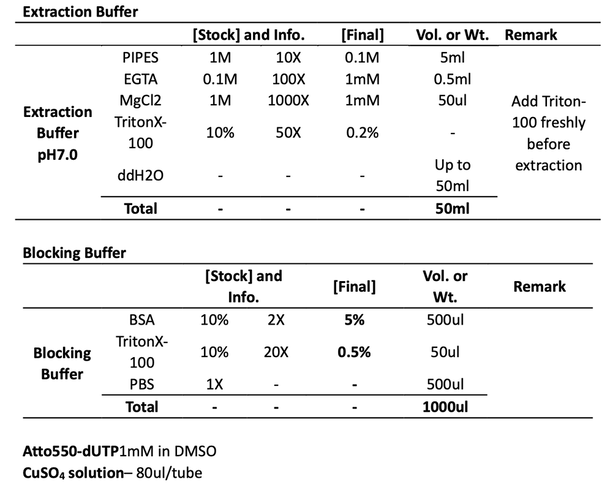
Transfer the cell culture dish to B220. Wash cells with PBS briefly. Incubate the cells with Extraction Buffer for 1min to extract the dissociative atto550-dUTP and EdU in the cytoplasm. Rinse once with PBS before the 10min fixation by 4% PFA and 0.1% GA in PBS, followed by 5min incubation of 1mg/ml NaBH4 quenching. Wash with PBS and keep in PBS at 4C.
Note: DO NOT QUENCH !!!
Day 3 - Wed May 07 2014
18:00 To permeabilize the cells, incubate the cells with Blocking buffer for 30min at RT
18:30 Remove the Blocking buffer, then wash the cells in each well twice with 1 mL of 5% BSA in PBS. Remove the wash solution.
Day 0 – Sun May 04 201
16:00 thaw and passage Hela cells from liquid nitrogen.
Day 1 – Mon May 05 2014 11:00 passage cell
In 6‐well plate, passage the Hela cells 1:2 in fresh medium. 19:00 1st Block (15hrs)
In 6‐well plate, change fresh medium.
Add Thymidine to final concentration 2mM to Hela cells medium to block the cells.
Day 2 – Tue May 06 2014 10:00 1st Release (10hrs)
To release the cells from S phase, wash cells with 1X DPBS thrice to remove the high concentration of Thymidine, passage Hela cells to coverslip bottomed petri dish to final density 20% ~ 30%.
#VII‐1 #VII‐2 #VII‐3 Hela cells; plate some cells in 6well plate as #VII‐4 dish;
20:00 2nd Block (15hrs)
In 6‐well plate and 3 coverslip bottomed petri dishes, change fresh medium.
Add Aphidicolin to final concentration 2ug/ml to Hela cell medium
Day 3 - Wed May 07 2014
10:20 make 12ml fresh medium containing 10uM EdU in 15ml tube.
Early-S # VI-1dish Mid-S # VI-2 dish Late-S # VI-3dish
10:30 13:00 15:30
100ul PBS + 25ul FuGene6, incubate 5min at RT. Add 1ul 1mM Atto550-dUTP (Jena Bioscience) to the mix, incubate 20min at 4C freezer, final concentration of the Atto550-dUTP is ~10uM.
11:00 # VI-1,# VI-2,# VI-3, # VI-4 dish To release the cells, wash cells with PBS thrice to remove the aphidicolin, change to 4ml fresh medium (DMEM), then the cells will enter S phase.
Whole Chrom # V-4 dish
11:00 3ml fresh medium + 1ml fresh medium with 10uM EdU. Final EdU is 2.5uM.
Early-S # VI-1dish Mid-S # VI-2 dish Late-S # VI-3dish
11:00 13:30 16:00
Wash cells with PBS twice. Add the ~100ul Atto550-dUTP-FuGene6-PBS Mix to PBS washed cells for 10min at RT. Note: be careful, do not let the cell dry!!!
11:10 13:40 16:10
Fill the dish with 4ml FRESH DMEM. Culture cells at 37C for 0.5 hrs.
11:40 14:10 16:40
4ml DMEM medium with EdU, the concentration is 10uM.
12:10 14:40 17:10
Transfer the cell culture dish to B220. Wash cells with PBS briefly. Incubate the cells with Extraction Buffer for 1min to extract the dissociative atto550-dUTP and EdU in the cytoplasm. Rinse once with PBS before the 10min fixation by 4% PFA and 0.1% GA in PBS, followed by 5min incubation of 1mg/ml NaBH4 quenching. Wash with PBS and keep in PBS at 4C.
Note: DO NOT QUENCH !!!
Day 3 - Wed May 07 2014
18:00 To permeabilize the cells, incubate the cells with Blocking buffer for 30min at RT
18:30 Remove the Blocking buffer, then wash the cells in each well twice with 1 mL of 5% BSA in PBS. Remove the wash solution.
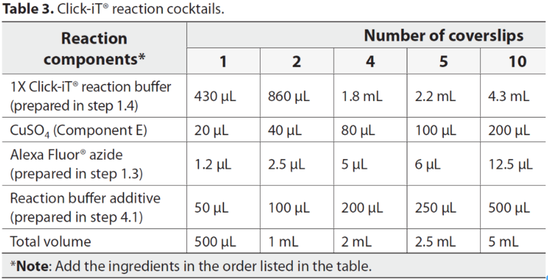
18:30 Add 0.5 mL of Click-iT® reaction cocktail to each well containing a coverslip.
Rock theplate briefly to insure that the reaction cocktail is distributed evenly over the coverslip. Incubate the plate for 30 minutes at room temperature, protected from light.
19:00 Remove the reaction cocktail, then wash each well once with 1 mL of 5% BSA in PBS. Remove the wash solution.
Rock theplate briefly to insure that the reaction cocktail is distributed evenly over the coverslip. Incubate the plate for 30 minutes at room temperature, protected from light.
19:00 Remove the reaction cocktail, then wash each well once with 1 mL of 5% BSA in PBS. Remove the wash solution.
- blocking and permeabilization with 3% w/v bovine serum albumin (JacksonImmunoResearch Laboratories) and 0.5% v/v Triton X-100 in PBS for 30 min;
- Note: no need for an extra permeabilization
- staining for 60min with
- Hela – Mouse anti-LaminA/C
- washing with PBS;
- staining for 60 min with
- Hela – Mouse anti-LaminA/C – Donkey anti Mouse Atto488
- washing with PBS;
- post-fixation in a mixture of 3%formaldehyde and 0.1% glutaraldehyde in PBS for 10 min;
- washing in PBS.