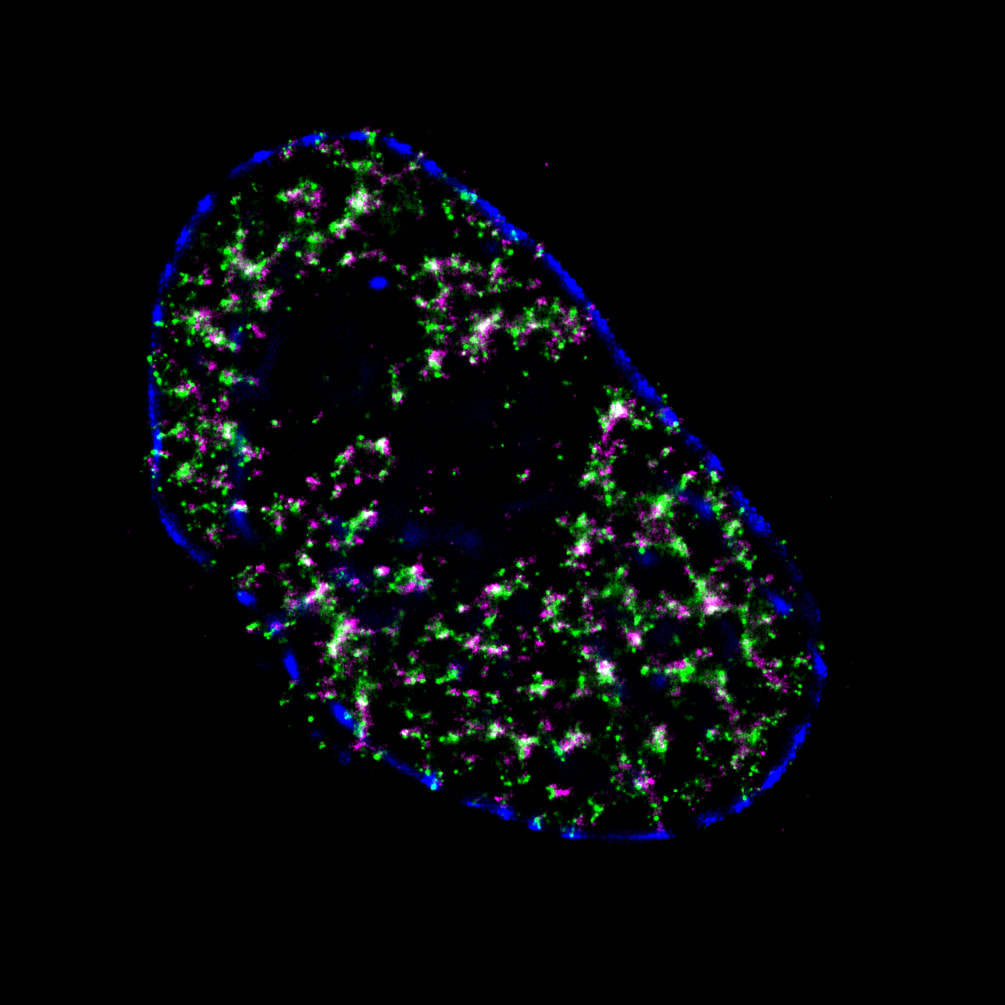
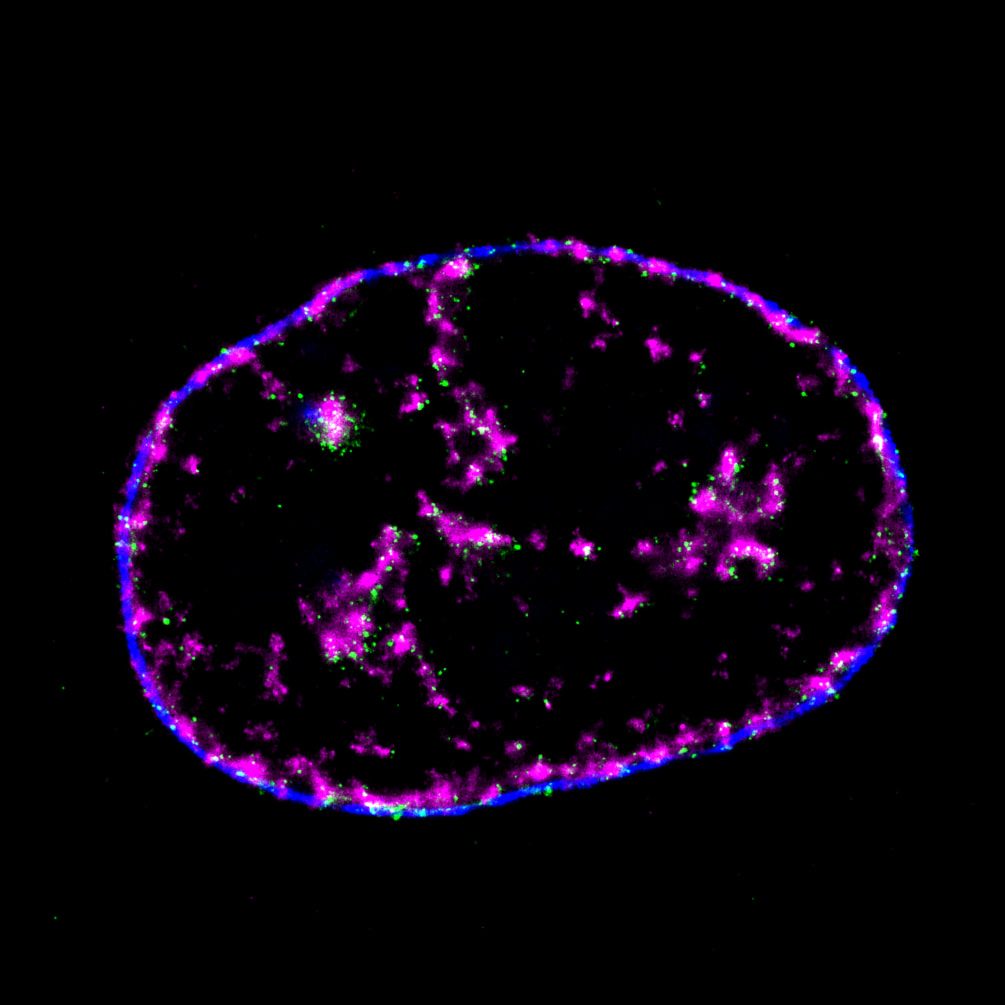
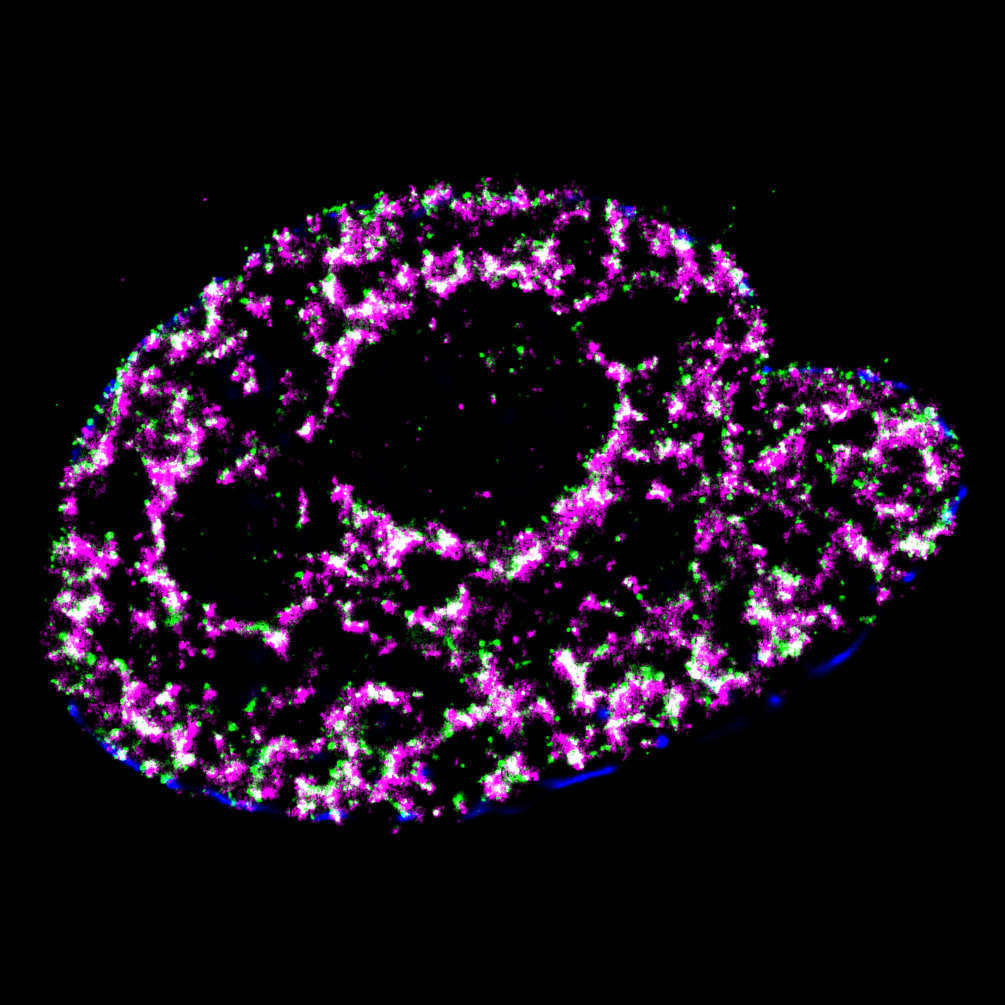
Pinpoint Chromatin 3D Structure and DNA Replication by Super-resolution Microscopy
Mammalian DNA replication is initiated at numerous replication origins, which are clustered into thousands of replication domains (RDs) across the genome. However, it remains unclear whether the replication origins within each RD are activated stochastically. To understand how replication is regulated at the sub-RD level, I have worked with Prof Xiaoliang Sunney XIE and Prof Xiaowei ZHUANG and their colleagues at Harvard and developed multi-colour 3D STORM chromatin imaging analysis. This new method advanced the field by allowing one to visualise the spatio-temporal organization, morphology, and in situ epigenetic signatures of individual replication foci (RFi) across S-phase. Importantly, I have revealed a hierarchical radial pattern of RFi propagation that reverses its directionality from early to late S-phase, and is diminished upon caffeine treatment or CTCF knockdown. Together with simulation and bioinformatics analyses, my research works point to a ‘CTCF-organized REplication Propagation’ (CoREP) model. The CoREP model suggests a non-random selection mechanism for replication activation mediated by CTCF at the sub-RD level, as well as the critical involvement of local chromatin environment in regulating replication in space and time [12].
The well-established super-resolution assay (optical configuration and analysis algorithms) with 30nm resolution in cell nucleus has also been applied to understand the role of PTEN protein in DNA repair [13], DNA oligo bio-conjugation and localization [14] and transcription factor in bacterial cells [15].
[12] Su, Qian Peter*#(equal contribution, corresponding author), W.Z. Zhao*, L. Meng, M. Ding, W. Zhang, Y. Gao, X.S. Xie#, and Y. Sun#. 2020. Super-resolution Imaging Reveals Spatio-temporal Propagation of Human Replication Foci Mediated by CTCF-organized Chromatin Structures. Proceedings of the National Academy of Sciences (PNAS). DOI: 10.1073/pnas.2001521117
[13] Wang, G.*, Y. Li*, P. Wang, H. Liang, M. Cui, M. Zhu, L. Guo, Qian Peter Su, Y. Sun, M.A. McNutt, and Y. Yin. 2015. PTEN regulates RPA1 and protects DNA replication forks. Cell Research (Cover) 25(11)
[14] Ren W., S. Wen, S.A. Tawfik, Qian Peter Su, G. Lin, L. Ju, M.J. Ford, H. Ghodke, A.M. van Oijen, D. Jin. 2018. Anisotropic functionalization of upconversion nanoparticles. Chemical Science 9(18).
[15] Liu, Z., D. Xing, Qian Peter Su, Y. Zhu, J. Zhang, et al Y. Sun. 2014. Super-resolution imaging and tracking of protein-protein interactions in sub-diffraction cellular space. Nature Comms 5: p. 4443.
The well-established super-resolution assay (optical configuration and analysis algorithms) with 30nm resolution in cell nucleus has also been applied to understand the role of PTEN protein in DNA repair [13], DNA oligo bio-conjugation and localization [14] and transcription factor in bacterial cells [15].
[12] Su, Qian Peter*#(equal contribution, corresponding author), W.Z. Zhao*, L. Meng, M. Ding, W. Zhang, Y. Gao, X.S. Xie#, and Y. Sun#. 2020. Super-resolution Imaging Reveals Spatio-temporal Propagation of Human Replication Foci Mediated by CTCF-organized Chromatin Structures. Proceedings of the National Academy of Sciences (PNAS). DOI: 10.1073/pnas.2001521117
[13] Wang, G.*, Y. Li*, P. Wang, H. Liang, M. Cui, M. Zhu, L. Guo, Qian Peter Su, Y. Sun, M.A. McNutt, and Y. Yin. 2015. PTEN regulates RPA1 and protects DNA replication forks. Cell Research (Cover) 25(11)
[14] Ren W., S. Wen, S.A. Tawfik, Qian Peter Su, G. Lin, L. Ju, M.J. Ford, H. Ghodke, A.M. van Oijen, D. Jin. 2018. Anisotropic functionalization of upconversion nanoparticles. Chemical Science 9(18).
[15] Liu, Z., D. Xing, Qian Peter Su, Y. Zhu, J. Zhang, et al Y. Sun. 2014. Super-resolution imaging and tracking of protein-protein interactions in sub-diffraction cellular space. Nature Comms 5: p. 4443.